使用 Scipy(Python)将经验分布与理论分布拟合?
- 2024-12-19 09:23:00
- admin 原创
- 141
问题描述:
简介:我有一个包含 30,000 多个整数值的列表,范围从 0 到 47(含 0 和 47),例如[0,0,0,0,..,1,1,1,1,...,2,2,2,2,...,47,47,47,...]从某个连续分布中采样。列表中的值不一定按顺序排列,但顺序对于这个问题来说并不重要。
问题:根据我的分布,我想计算任何给定值的 p 值(看到更大值的概率)。例如,正如您所见,0 的 p 值将接近 1,而更大数字的 p 值将趋向于 0。
我不知道我是否正确,但为了确定概率,我认为我需要将我的数据拟合到最适合描述我的数据的理论分布中。我假设需要某种拟合优度检验来确定最佳模型。
有没有办法用 Python (Scipy或Numpy) 实现这样的分析?你能举些例子吗?
解决方案 1:
利用平方误差和 (SSE) 进行分布拟合
这是对Saullo 的答案的更新和修改,它使用当前scipy.stats分布的完整列表,并返回分布的直方图和数据的直方图之间SSE最小的分布。
示例试穿
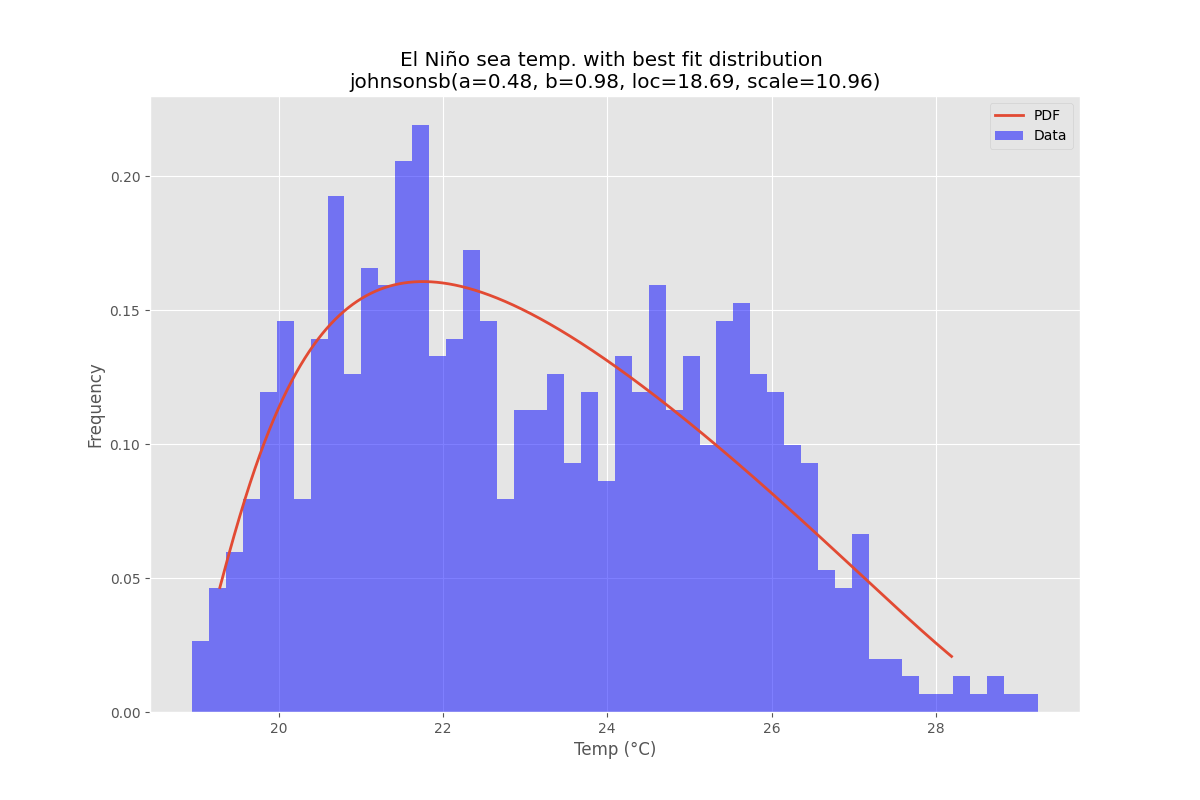
使用 的厄尔尼诺数据集statsmodels,拟合分布并确定误差。返回误差最小的分布。
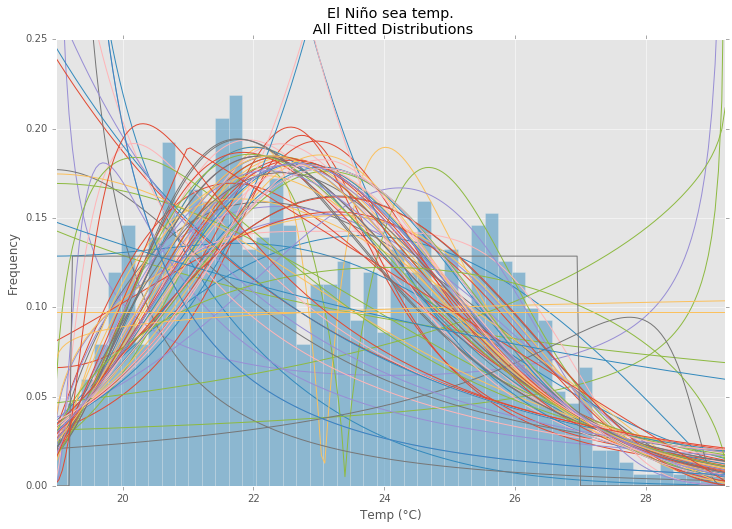
所有发行版
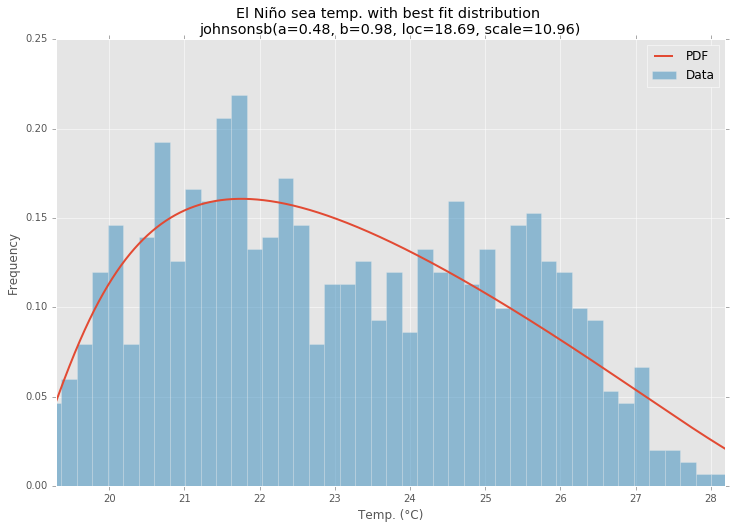
最佳拟合分布
示例代码
%matplotlib inline
import warnings
import numpy as np
import pandas as pd
import scipy.stats as st
import statsmodels.api as sm
from scipy.stats._continuous_distns import _distn_names
import matplotlib
import matplotlib.pyplot as plt
matplotlib.rcParams['figure.figsize'] = (16.0, 12.0)
matplotlib.style.use('ggplot')
# Create models from data
def best_fit_distribution(data, bins=200, ax=None):
"""Model data by finding best fit distribution to data"""
# Get histogram of original data
y, x = np.histogram(data, bins=bins, density=True)
x = (x + np.roll(x, -1))[:-1] / 2.0
# Best holders
best_distributions = []
# Estimate distribution parameters from data
for ii, distribution in enumerate([d for d in _distn_names if not d in ['levy_stable', 'studentized_range']]):
print("{:>3} / {:<3}: {}".format( ii+1, len(_distn_names), distribution ))
distribution = getattr(st, distribution)
# Try to fit the distribution
try:
# Ignore warnings from data that can't be fit
with warnings.catch_warnings():
warnings.filterwarnings('ignore')
# fit dist to data
params = distribution.fit(data)
# Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = distribution.pdf(x, loc=loc, scale=scale, *arg)
sse = np.sum(np.power(y - pdf, 2.0))
# if axis pass in add to plot
try:
if ax:
pd.Series(pdf, x).plot(ax=ax)
end
except Exception:
pass
# identify if this distribution is better
best_distributions.append((distribution, params, sse))
except Exception:
pass
return sorted(best_distributions, key=lambda x:x[2])
def make_pdf(dist, params, size=10000):
"""Generate distributions's Probability Distribution Function """
# Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
# Get sane start and end points of distribution
start = dist.ppf(0.01, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.01, loc=loc, scale=scale)
end = dist.ppf(0.99, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.99, loc=loc, scale=scale)
# Build PDF and turn into pandas Series
x = np.linspace(start, end, size)
y = dist.pdf(x, loc=loc, scale=scale, *arg)
pdf = pd.Series(y, x)
return pdf
# Load data from statsmodels datasets
data = pd.Series(sm.datasets.elnino.load_pandas().data.set_index('YEAR').values.ravel())
# Plot for comparison
plt.figure(figsize=(12,8))
ax = data.plot(kind='hist', bins=50, density=True, alpha=0.5, color=list(matplotlib.rcParams['axes.prop_cycle'])[1]['color'])
# Save plot limits
dataYLim = ax.get_ylim()
# Find best fit distribution
best_distibutions = best_fit_distribution(data, 200, ax)
best_dist = best_distibutions[0]
# Update plots
ax.set_ylim(dataYLim)
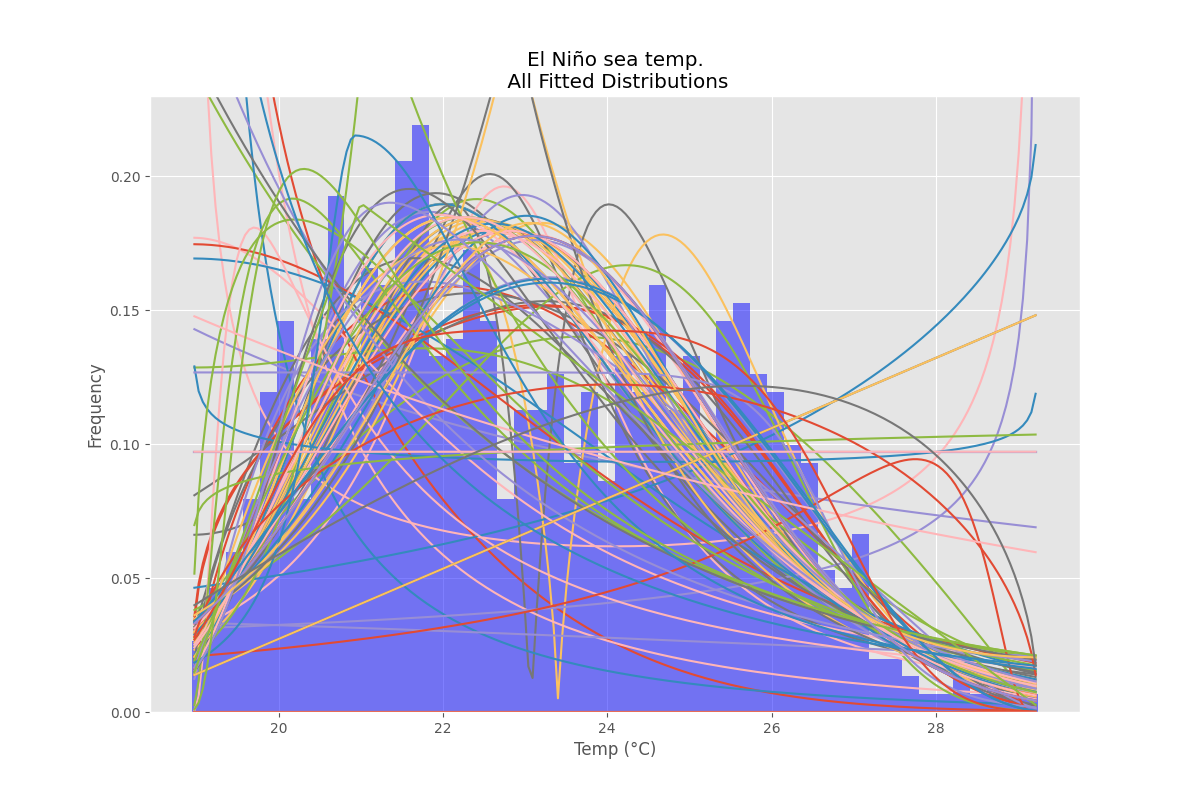
ax.set_title(u'El Niño sea temp.
All Fitted Distributions')
ax.set_xlabel(u'Temp (°C)')
ax.set_ylabel('Frequency')
# Make PDF with best params
pdf = make_pdf(best_dist[0], best_dist[1])
# Display
plt.figure(figsize=(12,8))
ax = pdf.plot(lw=2, label='PDF', legend=True)
data.plot(kind='hist', bins=50, density=True, alpha=0.5, label='Data', legend=True, ax=ax)
param_names = (best_dist[0].shapes + ', loc, scale').split(', ') if best_dist[0].shapes else ['loc', 'scale']
param_str = ', '.join(['{}={:0.2f}'.format(k,v) for k,v in zip(param_names, best_dist[1])])
dist_str = '{}({})'.format(best_dist[0].name, param_str)
ax.set_title(u'El Niño sea temp. with best fit distribution
' + dist_str)
ax.set_xlabel(u'Temp. (°C)')
ax.set_ylabel('Frequency')
解决方案 2:
SciPy v1.6.0 中实现了 90 多个分布函数。你可以使用它们的fit()方法测试其中一些函数如何适合你的数据。查看以下代码了解更多详细信息:
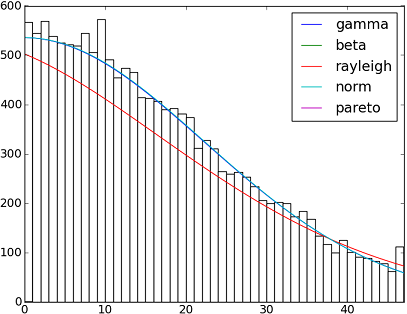
import matplotlib.pyplot as plt
import numpy as np
import scipy
import scipy.stats
size = 30000
x = np.arange(size)
y = scipy.int_(np.round_(scipy.stats.vonmises.rvs(5,size=size)*47))
h = plt.hist(y, bins=range(48))
dist_names = ['gamma', 'beta', 'rayleigh', 'norm', 'pareto']
for dist_name in dist_names:
dist = getattr(scipy.stats, dist_name)
params = dist.fit(y)
arg = params[:-2]
loc = params[-2]
scale = params[-1]
if arg:
pdf_fitted = dist.pdf(x, *arg, loc=loc, scale=scale) * size
else:
pdf_fitted = dist.pdf(x, loc=loc, scale=scale) * size
plt.plot(pdf_fitted, label=dist_name)
plt.xlim(0,47)
plt.legend(loc='upper right')
plt.show()
参考:
拟合分布、拟合优度、p 值。可以使用 Scipy (Python) 执行此操作吗?
使用 Scipy 进行分布拟合
这里列出了 Scipy 0.12.0 (VI) 中所有可用的分布函数的名称:
dist_names = [ 'alpha', 'anglit', 'arcsine', 'beta', 'betaprime', 'bradford', 'burr', 'cauchy', 'chi', 'chi2', 'cosine', 'dgamma', 'dweibull', 'erlang', 'expon', 'exponweib', 'exponpow', 'f', 'fatiguelife', 'fisk', 'foldcauchy', 'foldnorm', 'frechet_r', 'frechet_l', 'genlogistic', 'genpareto', 'genexpon', 'genextreme', 'gausshyper', 'gamma', 'gengamma', 'genhalflogistic', 'gilbrat', 'gompertz', 'gumbel_r', 'gumbel_l', 'halfcauchy', 'halflogistic', 'halfnorm', 'hypsecant', 'invgamma', 'invgauss', 'invweibull', 'johnsonsb', 'johnsonsu', 'ksone', 'kstwobign', 'laplace', 'logistic', 'loggamma', 'loglaplace', 'lognorm', 'lomax', 'maxwell', 'mielke', 'nakagami', 'ncx2', 'ncf', 'nct', 'norm', 'pareto', 'pearson3', 'powerlaw', 'powerlognorm', 'powernorm', 'rdist', 'reciprocal', 'rayleigh', 'rice', 'recipinvgauss', 'semicircular', 't', 'triang', 'truncexpon', 'truncnorm', 'tukeylambda', 'uniform', 'vonmises', 'wald', 'weibull_min', 'weibull_max', 'wrapcauchy']
解决方案 3:
你可以尝试distfit 库。如果你有更多问题,请告诉我,我也是这个开源库的开发者。
pip install distfit
# Create 1000 random integers, value between [0-50]
X = np.random.randint(0, 50,1000)
# Retrieve P-value for y
y = [0,10,45,55,100]
# From the distfit library import the class distfit
from distfit import distfit
# Initialize.
# Set any properties here, such as alpha.
# The smoothing can be of use when working with integers. Otherwise your histogram
# may be jumping up-and-down, and getting the correct fit may be harder.
dist = distfit(alpha=0.05, smooth=10)
# Search for best theoretical fit on your empirical data
dist.fit_transform(X)
> [distfit] >fit..
> [distfit] >transform..
> [distfit] >[norm ] [RSS: 0.0037894] [loc=23.535 scale=14.450]
> [distfit] >[expon ] [RSS: 0.0055534] [loc=0.000 scale=23.535]
> [distfit] >[pareto ] [RSS: 0.0056828] [loc=-384473077.778 scale=384473077.778]
> [distfit] >[dweibull ] [RSS: 0.0038202] [loc=24.535 scale=13.936]
> [distfit] >[t ] [RSS: 0.0037896] [loc=23.535 scale=14.450]
> [distfit] >[genextreme] [RSS: 0.0036185] [loc=18.890 scale=14.506]
> [distfit] >[gamma ] [RSS: 0.0037600] [loc=-175.505 scale=1.044]
> [distfit] >[lognorm ] [RSS: 0.0642364] [loc=-0.000 scale=1.802]
> [distfit] >[beta ] [RSS: 0.0021885] [loc=-3.981 scale=52.981]
> [distfit] >[uniform ] [RSS: 0.0012349] [loc=0.000 scale=49.000]
# Best fitted model
best_distr = dist.model
print(best_distr)
# Uniform shows best fit, with 95% CII (confidence intervals), and all other parameters
> {'distr': <scipy.stats._continuous_distns.uniform_gen at 0x16de3a53160>,
> 'params': (0.0, 49.0),
> 'name': 'uniform',
> 'RSS': 0.0012349021241149533,
> 'loc': 0.0,
> 'scale': 49.0,
> 'arg': (),
> 'CII_min_alpha': 2.45,
> 'CII_max_alpha': 46.55}
# Ranking distributions
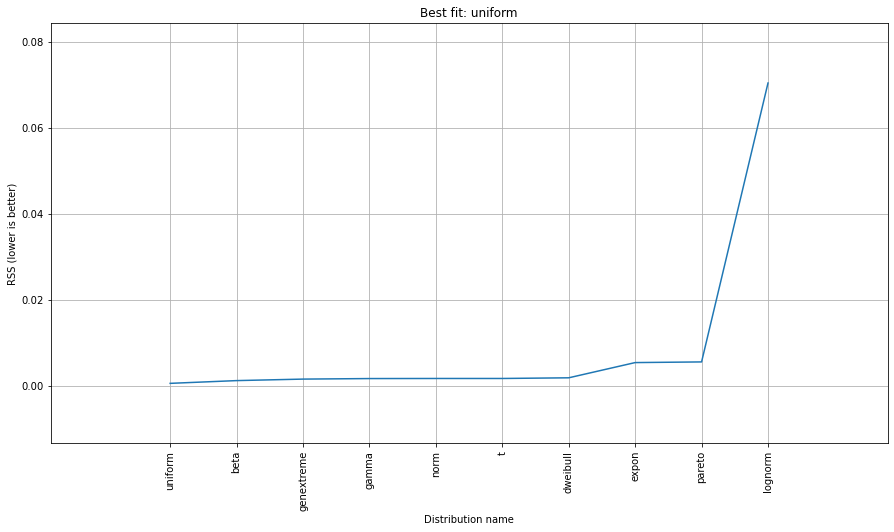
dist.summary
# Plot the summary of fitted distributions
dist.plot_summary()
# Make prediction on new datapoints based on the fit
dist.predict(y)
# Retrieve your pvalues with
dist.y_pred
# array(['down', 'none', 'none', 'up', 'up'], dtype='<U4')
dist.y_proba
array([0.02040816, 0.02040816, 0.02040816, 0. , 0. ])
# Or in one dataframe
dist.df
# The plot function will now also include the predictions of y
dist.plot()
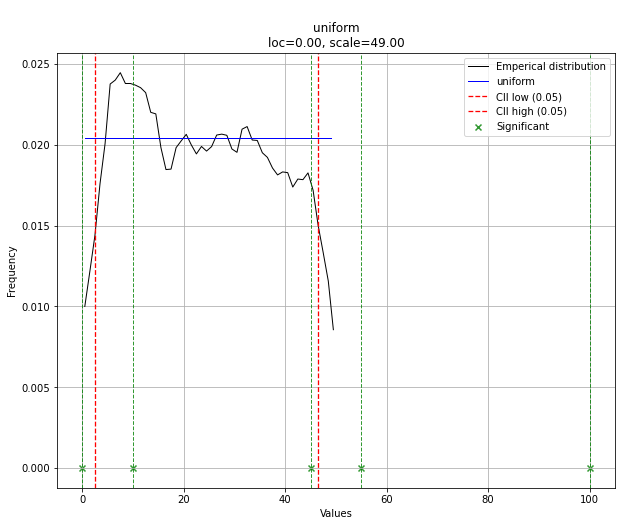
请注意,在这种情况下,由于分布均匀,所有点都将具有显著性。如有必要,您可以使用 dist.y_pred 进行过滤。
更详细的信息和示例可以在文档页面中找到。
解决方案 4:
fit()@Saullo Castro 提到的方法提供了最大似然估计 (MLE)。最适合您数据的分布是给您最高值的分布,可以通过几种不同的方式确定:
1,给出最高对数似然的那个。
2、给出最小 AIC、BIC 或 BICc 值的那个(参见 wiki: http: //en.wikipedia.org/wiki/Akaike_information_criterion,基本上可以看作是调整参数数量的对数似然,因为具有更多参数的分布预计会更好地拟合)
3、最大化贝叶斯后验概率。(参见维基: http: //en.wikipedia.org/wiki/Posterior_probability)
当然,如果您已经有一个可以描述您的数据的分布(基于您所在特定领域的理论)并且想要坚持使用这个分布,那么您将跳过识别最佳拟合分布的步骤。
scipy没有附带计算对数似然的函数(尽管提供了 MLE 方法),但硬编码很容易:请参阅scipy.stat.distributions的内置概率密度函数是否比用户提供的函数慢?
解决方案 5:
AFAICU,您的分布是离散的(而且只是离散的)。因此,只需计算不同值的频率并对其进行归一化就足以满足您的目的。因此,举一个例子来证明这一点:
In []: values= [0, 0, 0, 0, 0, 1, 1, 1, 1, 2, 2, 2, 3, 3, 4]
In []: counts= asarray(bincount(values), dtype= float)
In []: cdf= counts.cumsum()/ counts.sum()
因此,看到高于的值的概率1很简单(根据互补累积分布函数(ccdf):
In []: 1- cdf[1]
Out[]: 0.40000000000000002
请注意,ccdf与生存函数 (sf)密切相关,但它也是用离散分布定义的,而sf仅为连续分布定义。
解决方案 6:
以下代码是通用答案的版本,但经过了修正和澄清。
import numpy as np
import pandas as pd
import scipy.stats as st
import statsmodels.api as sm
import matplotlib as mpl
import matplotlib.pyplot as plt
import math
import random
mpl.style.use("ggplot")
def danoes_formula(data):
"""
DANOE'S FORMULA
https://en.wikipedia.org/wiki/Histogram#Doane's_formula
"""
N = len(data)
skewness = st.skew(data)
sigma_g1 = math.sqrt((6*(N-2))/((N+1)*(N+3)))
num_bins = 1 + math.log(N,2) + math.log(1+abs(skewness)/sigma_g1,2)
num_bins = round(num_bins)
return num_bins
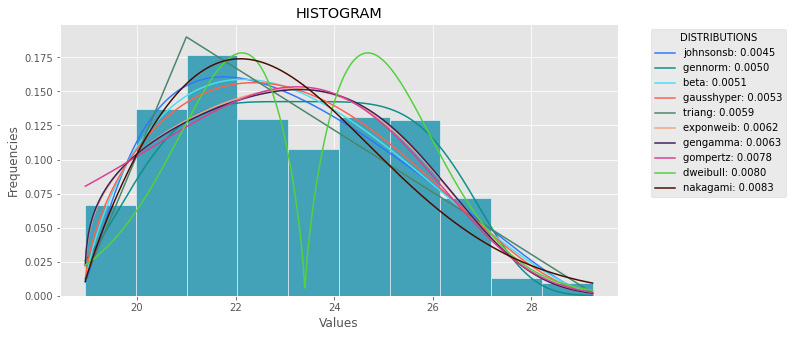
def plot_histogram(data, results, n):
## n first distribution of the ranking
N_DISTRIBUTIONS = {k: results[k] for k in list(results)[:n]}
## Histogram of data
plt.figure(figsize=(10, 5))
plt.hist(data, density=True, ec='white', color=(63/235, 149/235, 170/235))
plt.title('HISTOGRAM')
plt.xlabel('Values')
plt.ylabel('Frequencies')
## Plot n distributions
for distribution, result in N_DISTRIBUTIONS.items():
# print(i, distribution)
sse = result[0]
arg = result[1]
loc = result[2]
scale = result[3]
x_plot = np.linspace(min(data), max(data), 1000)
y_plot = distribution.pdf(x_plot, loc=loc, scale=scale, *arg)
plt.plot(x_plot, y_plot, label=str(distribution)[32:-34] + ": " + str(sse)[0:6], color=(random.uniform(0, 1), random.uniform(0, 1), random.uniform(0, 1)))
plt.legend(title='DISTRIBUTIONS', bbox_to_anchor=(1.05, 1), loc='upper left')
plt.show()
def fit_data(data):
## st.frechet_r,st.frechet_l: are disbled in current SciPy version
## st.levy_stable: a lot of time of estimation parameters
ALL_DISTRIBUTIONS = [
st.alpha,st.anglit,st.arcsine,st.beta,st.betaprime,st.bradford,st.burr,st.cauchy,st.chi,st.chi2,st.cosine,
st.dgamma,st.dweibull,st.erlang,st.expon,st.exponnorm,st.exponweib,st.exponpow,st.f,st.fatiguelife,st.fisk,
st.foldcauchy,st.foldnorm, st.genlogistic,st.genpareto,st.gennorm,st.genexpon,
st.genextreme,st.gausshyper,st.gamma,st.gengamma,st.genhalflogistic,st.gilbrat,st.gompertz,st.gumbel_r,
st.gumbel_l,st.halfcauchy,st.halflogistic,st.halfnorm,st.halfgennorm,st.hypsecant,st.invgamma,st.invgauss,
st.invweibull,st.johnsonsb,st.johnsonsu,st.ksone,st.kstwobign,st.laplace,st.levy,st.levy_l,
st.logistic,st.loggamma,st.loglaplace,st.lognorm,st.lomax,st.maxwell,st.mielke,st.nakagami,st.ncx2,st.ncf,
st.nct,st.norm,st.pareto,st.pearson3,st.powerlaw,st.powerlognorm,st.powernorm,st.rdist,st.reciprocal,
st.rayleigh,st.rice,st.recipinvgauss,st.semicircular,st.t,st.triang,st.truncexpon,st.truncnorm,st.tukeylambda,
st.uniform,st.vonmises,st.vonmises_line,st.wald,st.weibull_min,st.weibull_max,st.wrapcauchy
]
MY_DISTRIBUTIONS = [st.beta, st.expon, st.norm, st.uniform, st.johnsonsb, st.gennorm, st.gausshyper]
## Calculae Histogram
num_bins = danoes_formula(data)
frequencies, bin_edges = np.histogram(data, num_bins, density=True)
central_values = [(bin_edges[i] + bin_edges[i+1])/2 for i in range(len(bin_edges)-1)]
results = {}
for distribution in MY_DISTRIBUTIONS:
## Get parameters of distribution
params = distribution.fit(data)
## Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
## Calculate fitted PDF and error with fit in distribution
pdf_values = [distribution.pdf(c, loc=loc, scale=scale, *arg) for c in central_values]
## Calculate SSE (sum of squared estimate of errors)
sse = np.sum(np.power(frequencies - pdf_values, 2.0))
## Build results and sort by sse
results[distribution] = [sse, arg, loc, scale]
results = {k: results[k] for k in sorted(results, key=results.get)}
return results
def main():
## Import data
data = pd.Series(sm.datasets.elnino.load_pandas().data.set_index('YEAR').values.ravel())
results = fit_data(data)
plot_histogram(data, results, 5)
if __name__ == "__main__":
main()
如果你想做更详细的分析,我推荐Phittter库
import phitter
data: list[int | float] = [...]
phitter_cont = phitter.PHITTER(
data=data,
fit_type="continuous",
num_bins=15,
confidence_level=0.95,
minimum_sse=1e-2,
distributions_to_fit=["beta", "normal", "fatigue_life", "triangular"],
)
phitter_cont.fit(n_workers=6)
解决方案 7:
虽然上述许多答案都是完全有效的,但似乎没有人能完全回答你的问题,特别是以下部分:
我不知道我是否正确,但为了确定概率,我认为我需要将我的数据拟合到最适合描述我的数据的理论分布中。我假设需要某种拟合优度检验来确定最佳模型。
参数化方法
这是您所描述的使用某些理论分布并将参数拟合到您的数据的过程,并且有一些关于如何做到这一点的很好的答案。
非参数方法
但是,您也可以使用非参数方法来解决您的问题,这意味着您根本不假设任何底层分布。
通过使用所谓的经验分布函数,其等于:
Fn(x)= SUM( I[X<=x] ) / n。因此,低于 x 的值的比例。
正如上述答案之一指出的那样,您感兴趣的是逆 CDF(累积分布函数),它等于1-F(x)
可以证明,经验分布函数将收敛到生成数据的任何“真实”CDF。
此外,可以通过以下方法直接构建 1-alpha 置信区间:
L(X) = max{Fn(x)-en, 0}
U(X) = min{Fn(x)+en, 0}
en = sqrt( (1/2n)*log(2/alpha)
然后对于所有 x,P( L(X) <= F(X) <= U(X) ) >= 1-alpha 。
我很惊讶PierrOz 的答案有 0 票,而它是使用非参数方法估计 F(x) 的问题的完全有效的答案。
请注意,您提到的 P(X>=x)=0(对于任何 x>47)的问题只是个人偏好,可能会导致您选择参数方法而不是非参数方法。然而,这两种方法都是您问题的完全有效的解决方案。
有关上述陈述的更多细节和证明,我建议你看一下 Larry A. Wasserman 的《统计学全书:统计推断简明课程》。这是一本关于参数和非参数推断的优秀书籍。
编辑:
由于您特别要求提供一些 python 示例,因此可以使用 numpy 来完成:
import numpy as np
def empirical_cdf(data, x):
return np.sum(x<=data)/len(data)
def p_value(data, x):
return 1-empirical_cdf(data, x)
# Generate some data for demonstration purposes
data = np.floor(np.random.uniform(low=0, high=48, size=30000))
print(empirical_cdf(data, 20))
print(p_value(data, 20)) # This is the value you're interested in
解决方案 8:
我发现最简单的方法是使用 fitter 模块,您pip install fitter只需导入 pandas 数据集即可。它具有内置函数,可以从 scipy 搜索所有 80 个分布,并通过各种方法获得与数据的最佳拟合。示例:
f = Fitter(height, distributions=['gamma','lognorm', "beta","burr","norm"])
f.fit()
f.summary()
这里作者提供了一个发行版列表,因为扫描全部 80 个发行版可能会很耗时。
f.get_best(method = 'sumsquare_error')
这将为你提供 5 个符合其拟合标准的最佳分布:
sumsquare_error aic bic kl_div
chi2 0.000010 1716.234916 -1945.821606 inf
gamma 0.000010 1716.234909 -1945.821606 inf
rayleigh 0.000010 1711.807360 -1945.526026 inf
norm 0.000011 1758.797036 -1934.865211 inf
cauchy 0.000011 1762.735606 -1934.803414 inf
您还拥有distributions=get_common_distributions()包含大约 10 个最常用分布的属性,并且可以为您进行拟合和检查。
它还具有许多其他功能,如绘制直方图,所有完整的文档可以在这里找到。
对于科学家、工程师以及一般人来说,这是一个被严重低估的模块。
解决方案 9:
对我来说这听起来像是概率密度估计问题。
from scipy.stats import gaussian_kde
occurences = [0,0,0,0,..,1,1,1,1,...,2,2,2,2,...,47]
values = range(0,48)
kde = gaussian_kde(map(float, occurences))
p = kde(values)
p = p/sum(p)
print "P(x>=1) = %f" % sum(p[1:])
另请参阅http://jpktd.blogspot.com/2009/03/using-gaussian-kernel-density.html。
解决方案 10:
将数据存储在字典中怎么样?其中的键是 0 到 47 之间的数字,值是原始列表中相关键出现的次数。
因此,您的可能性 p(x) 将是所有大于 x 的键值的总和除以 30000。
解决方案 11:
使用OpenTURNS时,我会使用 BIC 标准来选择最适合此类数据的分布。这是因为该标准不会给具有更多参数的分布带来太大优势。事实上,如果分布具有更多参数,则拟合分布更容易接近数据。此外,在这种情况下,Kolmogorov-Smirnov 可能没有意义,因为测量值中的小误差会对 p 值产生巨大影响。
为了说明该过程,我加载了厄尔尼诺数据,其中包含 1950 年至 2010 年的 732 个月温度测量数据:
import statsmodels.api as sm
dta = sm.datasets.elnino.load_pandas().data
dta['YEAR'] = dta.YEAR.astype(int).astype(str)
dta = dta.set_index('YEAR').T.unstack()
data = dta.values
使用静态方法很容易获得 30 个内置的单变量分布工厂GetContinuousUniVariateFactories。完成后,BestModelBIC静态方法将返回最佳模型和相应的 BIC 分数。
sample = ot.Sample([[p] for p in data]) # data reshaping
tested_factories = ot.DistributionFactory.GetContinuousUniVariateFactories()
best_model, best_bic = ot.FittingTest.BestModelBIC(sample,
tested_factories)
print("Best=",best_model)
打印结果为:
Best= Beta(alpha = 1.64258, beta = 2.4348, a = 18.936, b = 29.254)
为了以图形方式比较与直方图的拟合度,我使用了drawPDF最佳分布的方法。
import openturns.viewer as otv
graph = ot.HistogramFactory().build(sample).drawPDF()
bestPDF = best_model.drawPDF()
bestPDF.setColors(["blue"])
graph.add(bestPDF)
graph.setTitle("Best BIC fit")
name = best_model.getImplementation().getClassName()
graph.setLegends(["Histogram",name])
graph.setXTitle("Temperature (°C)")
otv.View(graph)
得出的结果为:
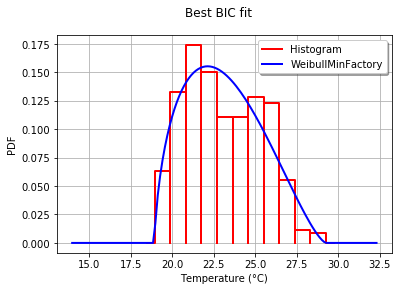
BestModelBIC文档中提供了有关此主题的更多详细信息。可以将 Scipy 分布包含在SciPyDistribution中,甚至可以将 ChaosPy 分布包含在ChaosPyDistribution中,但我想当前脚本可以满足大多数实际目的。
解决方案 12:
我从第一个答案重新设计了分布函数,其中包括一个选择参数,用于选择拟合优度检验之一,这将缩小适合数据的分布函数范围:
import numpy as np
import pandas as pd
import scipy.stats as st
import matplotlib.pyplot as plt
import pylab
def make_hist(data):
#### General code:
bins_formulas = ['auto', 'fd', 'scott', 'rice', 'sturges', 'doane', 'sqrt']
bins = np.histogram_bin_edges(a=data, bins='fd', range=(min(data), max(data)))
# print('Bin value = ', bins)
# Obtaining the histogram of data:
Hist, bin_edges = histogram(a=data, bins=bins, range=(min(data), max(data)), density=True)
bin_mid = (bin_edges + np.roll(bin_edges, -1))[:-1] / 2.0 # go from bin edges to bin middles
return Hist, bin_mid
def make_pdf(dist, params, size):
"""Generate distributions's Probability Distribution Function """
# Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
# Get sane start and end points of distribution
start = dist.ppf(0.01, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.01, loc=loc, scale=scale)
end = dist.ppf(0.99, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.99, loc=loc, scale=scale)
# Build PDF and turn into pandas Series
x = np.linspace(start, end, size)
y = dist.pdf(x, loc=loc, scale=scale, *arg)
pdf = pd.Series(y, x)
return pdf, x, y
def compute_r2_test(y_true, y_predicted):
sse = sum((y_true - y_predicted)**2)
tse = (len(y_true) - 1) * np.var(y_true, ddof=1)
r2_score = 1 - (sse / tse)
return r2_score, sse, tse
def get_best_distribution_2(data, method, plot=False):
dist_names = ['alpha', 'anglit', 'arcsine', 'beta', 'betaprime', 'bradford', 'burr', 'cauchy', 'chi', 'chi2', 'cosine', 'dgamma', 'dweibull', 'erlang', 'expon', 'exponweib', 'exponpow', 'f', 'fatiguelife', 'fisk', 'foldcauchy', 'foldnorm', 'frechet_r', 'frechet_l', 'genlogistic', 'genpareto', 'genexpon', 'genextreme', 'gausshyper', 'gamma', 'gengamma', 'genhalflogistic', 'gilbrat', 'gompertz', 'gumbel_r', 'gumbel_l', 'halfcauchy', 'halflogistic', 'halfnorm', 'hypsecant', 'invgamma', 'invgauss', 'invweibull', 'johnsonsb', 'johnsonsu', 'ksone', 'kstwobign', 'laplace', 'logistic', 'loggamma', 'loglaplace', 'lognorm', 'lomax', 'maxwell', 'mielke', 'moyal', 'nakagami', 'ncx2', 'ncf', 'nct', 'norm', 'pareto', 'pearson3', 'powerlaw', 'powerlognorm', 'powernorm', 'rdist', 'reciprocal', 'rayleigh', 'rice', 'recipinvgauss', 'semicircular', 't', 'triang', 'truncexpon', 'truncnorm', 'tukeylambda', 'uniform', 'vonmises', 'wald', 'weibull_min', 'weibull_max', 'wrapcauchy']
# Applying the Goodness-to-fit tests to select the best distribution that fits the data:
dist_results = []
dist_IC_results = []
params = {}
params_IC = {}
params_SSE = {}
if method == 'sse':
########################################################################################################################
######################################## Sum of Square Error (SSE) test ################################################
########################################################################################################################
# Best holders
best_distribution = st.norm
best_params = (0.0, 1.0)
best_sse = np.inf
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
N_len = len(list(data))
# Obtaining the histogram:
Hist_data, bin_data = make_hist(data=data)
# fit dist to data
params_dist = dist.fit(data)
# Separate parts of parameters
arg = params_dist[:-2]
loc = params_dist[-2]
scale = params_dist[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = dist.pdf(bin_data, loc=loc, scale=scale, *arg)
sse = np.sum(np.power(Hist_data - pdf, 2.0))
# identify if this distribution is better
if best_sse > sse > 0:
best_distribution = dist
best_params = params_dist
best_stat_test_val = sse
print('
################################ Sum of Square Error test parameters #####################################')
best_dist = best_distribution
print("Best fitting distribution (SSE test) :" + str(best_dist))
print("Best SSE value (SSE test) :" + str(best_stat_test_val))
print("Parameters for the best fit (SSE test) :" + str(params[best_dist]))
print('###########################################################################################################
')
########################################################################################################################
########################################################################################################################
########################################################################################################################
if method == 'r2':
########################################################################################################################
##################################################### R Square (R^2) test ##############################################
########################################################################################################################
# Best holders
best_distribution = st.norm
best_params = (0.0, 1.0)
best_r2 = np.inf
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
N_len = len(list(data))
# Obtaining the histogram:
Hist_data, bin_data = make_hist(data=data)
# fit dist to data
params_dist = dist.fit(data)
# Separate parts of parameters
arg = params_dist[:-2]
loc = params_dist[-2]
scale = params_dist[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = dist.pdf(bin_data, loc=loc, scale=scale, *arg)
r2 = compute_r2_test(y_true=Hist_data, y_predicted=pdf)
# identify if this distribution is better
if best_r2 > r2 > 0:
best_distribution = dist
best_params = params_dist
best_stat_test_val = r2
print('
############################## R Square test parameters ###########################################')
best_dist = best_distribution
print("Best fitting distribution (R^2 test) :" + str(best_dist))
print("Best R^2 value (R^2 test) :" + str(best_stat_test_val))
print("Parameters for the best fit (R^2 test) :" + str(params[best_dist]))
print('#####################################################################################################
')
########################################################################################################################
########################################################################################################################
########################################################################################################################
if method == 'ic':
########################################################################################################################
######################################## Information Criteria (IC) test ################################################
########################################################################################################################
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
N_len = len(list(data))
# Obtaining the histogram:
Hist_data, bin_data = make_hist(data=data)
# fit dist to data
params_dist = dist.fit(data)
# Separate parts of parameters
arg = params_dist[:-2]
loc = params_dist[-2]
scale = params_dist[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = dist.pdf(bin_data, loc=loc, scale=scale, *arg)
sse = np.sum(np.power(Hist_data - pdf, 2.0))
# Obtaining the log of the pdf:
loglik = np.sum(dist.logpdf(bin_data, *params_dist))
k = len(params_dist[:])
n = len(data)
aic = 2 * k - 2 * loglik
bic = n * np.log(sse / n) + k * np.log(n)
dist_IC_results.append((dist_name, aic))
# dist_IC_results.append((dist_name, bic))
# select the best fitted distribution and store the name of the best fit and its IC value
best_dist, best_ic = (min(dist_IC_results, key=lambda item: item[1]))
print('
############################ Information Criteria (IC) test parameters ##################################')
print("Best fitting distribution (IC test) :" + str(best_dist))
print("Best IC value (IC test) :" + str(best_ic))
print("Parameters for the best fit (IC test) :" + str(params[best_dist]))
print('###########################################################################################################
')
########################################################################################################################
########################################################################################################################
########################################################################################################################
if method == 'chi':
########################################################################################################################
################################################ Chi-Square (Chi^2) test ###############################################
########################################################################################################################
# Set up 50 bins for chi-square test
# Observed data will be approximately evenly distrubuted aross all bins
percentile_bins = np.linspace(0,100,51)
percentile_cutoffs = np.percentile(data, percentile_bins)
observed_frequency, bins = (np.histogram(data, bins=percentile_cutoffs))
cum_observed_frequency = np.cumsum(observed_frequency)
chi_square = []
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
# Obtaining the histogram:
Hist_data, bin_data = make_hist(data=data)
# fit dist to data
params_dist = dist.fit(data)
# Separate parts of parameters
arg = params_dist[:-2]
loc = params_dist[-2]
scale = params_dist[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = dist.pdf(bin_data, loc=loc, scale=scale, *arg)
# Get expected counts in percentile bins
# This is based on a 'cumulative distrubution function' (cdf)
cdf_fitted = dist.cdf(percentile_cutoffs, *arg, loc=loc, scale=scale)
expected_frequency = []
for bin in range(len(percentile_bins) - 1):
expected_cdf_area = cdf_fitted[bin + 1] - cdf_fitted[bin]
expected_frequency.append(expected_cdf_area)
# calculate chi-squared
expected_frequency = np.array(expected_frequency) * size
cum_expected_frequency = np.cumsum(expected_frequency)
ss = sum(((cum_expected_frequency - cum_observed_frequency) ** 2) / cum_observed_frequency)
chi_square.append(ss)
# Applying the Chi-Square test:
# D, p = scipy.stats.chisquare(f_obs=pdf, f_exp=Hist_data)
# print("Chi-Square test Statistics value for " + dist_name + " = " + str(D))
print("p value for " + dist_name + " = " + str(chi_square))
dist_results.append((dist_name, chi_square))
# select the best fitted distribution and store the name of the best fit and its p value
best_dist, best_stat_test_val = (min(dist_results, key=lambda item: item[1]))
print('
#################################### Chi-Square test parameters #######################################')
print("Best fitting distribution (Chi^2 test) :" + str(best_dist))
print("Best p value (Chi^2 test) :" + str(best_stat_test_val))
print("Parameters for the best fit (Chi^2 test) :" + str(params[best_dist]))
print('#########################################################################################################
')
########################################################################################################################
########################################################################################################################
########################################################################################################################
if method == 'ks':
########################################################################################################################
########################################## Kolmogorov-Smirnov (KS) test ################################################
########################################################################################################################
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
# Applying the Kolmogorov-Smirnov test:
D, p = st.kstest(data, dist_name, args=param)
# D, p = st.kstest(data, dist_name, args=param, N=N_len, alternative='greater')
# print("Kolmogorov-Smirnov test Statistics value for " + dist_name + " = " + str(D))
print("p value for " + dist_name + " = " + str(p))
dist_results.append((dist_name, p))
# select the best fitted distribution and store the name of the best fit and its p value
best_dist, best_stat_test_val = (max(dist_results, key=lambda item: item[1]))
print('
################################ Kolmogorov-Smirnov test parameters #####################################')
print("Best fitting distribution (KS test) :" + str(best_dist))
print("Best p value (KS test) :" + str(best_stat_test_val))
print("Parameters for the best fit (KS test) :" + str(params[best_dist]))
print('###########################################################################################################
')
########################################################################################################################
########################################################################################################################
########################################################################################################################
# Collate results and sort by goodness of fit (best at top)
results = pd.DataFrame()
results['Distribution'] = dist_names
results['chi_square'] = chi_square
# results['p_value'] = p_values
results.sort_values(['chi_square'], inplace=True)
# Plotting the distribution with histogram:
if plot:
bins_val = np.histogram_bin_edges(a=data, bins='fd', range=(min(data), max(data)))
plt.hist(x=data, bins=bins_val, range=(min(data), max(data)), density=True)
# pylab.hist(x=data, bins=bins_val, range=(min(data), max(data)))
best_param = params[best_dist]
best_dist_p = getattr(st, best_dist)
pdf, x_axis_pdf, y_axis_pdf = make_pdf(dist=best_dist_p, params=best_param, size=len(data))
plt.plot(x_axis_pdf, y_axis_pdf, color='red', label='Best dist ={0}'.format(best_dist))
plt.legend()
plt.title('Histogram and Distribution plot of data')
# plt.show()
plt.show(block=False)
plt.pause(5) # Pauses the program for 5 seconds
plt.close('all')
return best_dist, best_stat_test_val, params[best_dist]
然后继续 make_pdf 函数根据拟合优度检验获取所选分布。
解决方案 13:
根据Timothy Davenports 的回答,我重构了代码以用作库,并将其作为 github 和 pypi 项目提供,请参阅:
https://github.com/WolfgangFahl/pyProbabilityDistributionFit
https://github.com/WolfgangFahl/pyProbabilityDistributionFit/blob/main/pdffit/distfit.py
一个目标是使密度选项可用并将结果输出为文件。请参阅实现的主要部分:
bfd=BestFitDistribution(data)
for density in [True,False]:
suffix="density" if density else ""
bfd.analyze(title=u'El Niño sea temp.',x_label=u'Temp (°C)',y_label='Frequency',outputFilePrefix=f"/tmp/ElNinoPDF{suffix}",density=density)
# uncomment for interactive display
# plt.show()
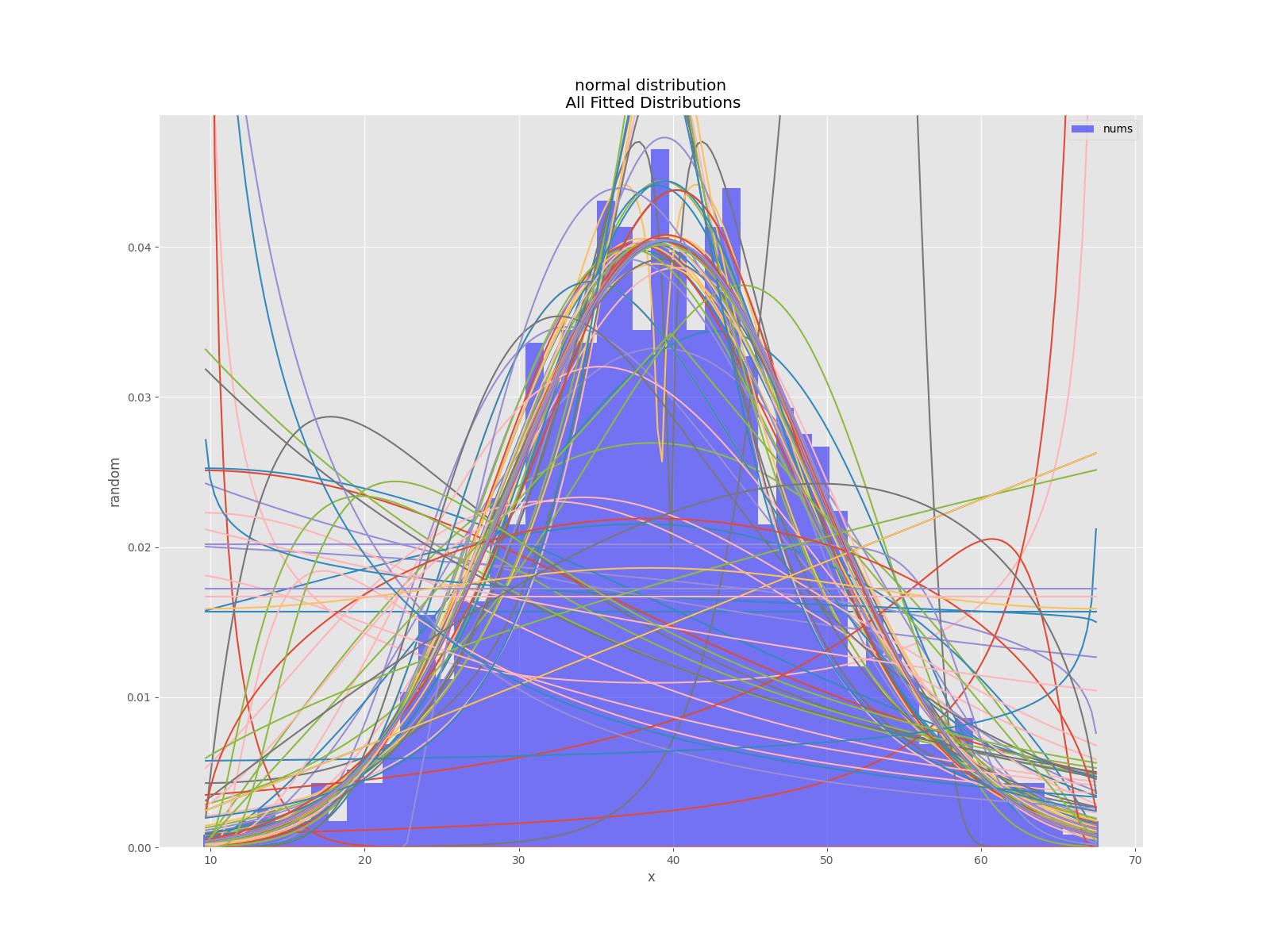
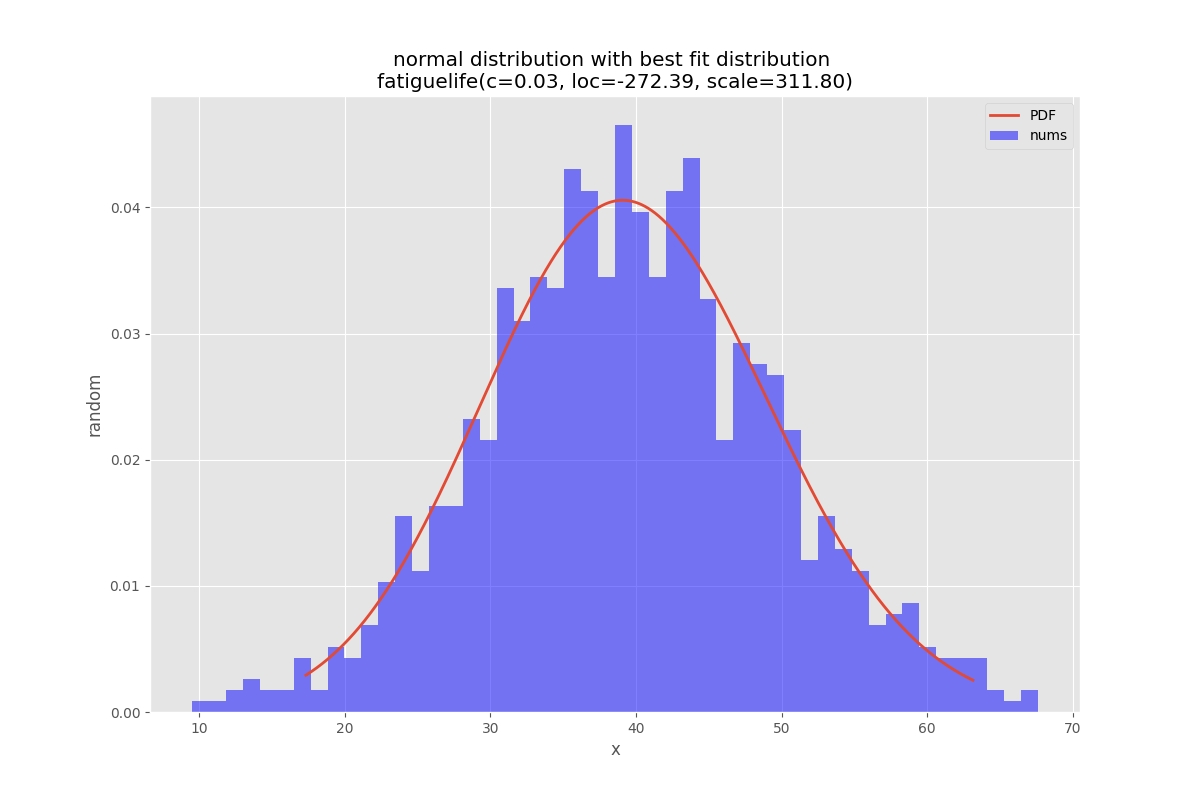
该库还有一个单元测试,例如
正态分布测试
def testNormal(self):
'''
test the normal distribution
'''
# use euler constant as seed
np.random.seed(0)
# statistically relevant number of datapoints
datapoints=1000
a = np.random.normal(40, 10, datapoints)
df= pd.DataFrame({'nums':a})
outputFilePrefix="/tmp/normalDist"
bfd=BestFitDistribution(df,debug=True)
bfd.analyze(title="normal distribution",x_label="x",y_label="random",outputFilePrefix=outputFilePrefix)
 如果您发现问题或有改进空间,
如果您发现问题或有改进空间,
请将问题添加到项目中。讨论也已启用。
下面的代码可能不是最新的,请使用 pypi 或 github 存储库获取最新版本。
'''
Created on 2022-05-17
see
https://stackoverflow.com/questions/6620471/fitting-empirical-distribution-to-theoretical-ones-with-scipy-python/37616966#37616966
@author: https://stackoverflow.com/users/832621/saullo-g-p-castro
see https://stackoverflow.com/a/37616966/1497139
@author: https://stackoverflow.com/users/2087463/tmthydvnprt
see https://stackoverflow.com/a/37616966/1497139
@author: https://stackoverflow.com/users/1497139/wolfgang-fahl
see
'''
import traceback
import sys
import warnings
import numpy as np
import pandas as pd
import scipy.stats as st
from scipy.stats._continuous_distns import _distn_names
import statsmodels.api as sm
import matplotlib
import matplotlib.pyplot as plt
class BestFitDistribution():
'''
Find the best Probability Distribution Function for the given data
'''
def __init__(self,data,distributionNames:list=None,debug:bool=False):
'''
constructor
Args:
data(dataFrame): the data to analyze
distributionNames(list): list of distributionNames to try
debug(bool): if True show debugging information
'''
self.debug=debug
self.matplotLibParams()
if distributionNames is None:
self.distributionNames=[d for d in _distn_names if not d in ['levy_stable', 'studentized_range']]
else:
self.distributionNames=distributionNames
self.data=data
def matplotLibParams(self):
'''
set matplotlib parameters
'''
matplotlib.rcParams['figure.figsize'] = (16.0, 12.0)
#matplotlib.style.use('ggplot')
matplotlib.use("WebAgg")
# Create models from data
def best_fit_distribution(self,bins:int=200, ax=None,density:bool=True):
"""
Model data by finding best fit distribution to data
"""
# Get histogram of original data
y, x = np.histogram(self.data, bins=bins, density=density)
x = (x + np.roll(x, -1))[:-1] / 2.0
# Best holders
best_distributions = []
distributionCount=len(self.distributionNames)
# Estimate distribution parameters from data
for ii, distributionName in enumerate(self.distributionNames):
print(f"{ii+1:>3} / {distributionCount:<3}: {distributionName}")
distribution = getattr(st, distributionName)
# Try to fit the distribution
try:
# Ignore warnings from data that can't be fit
with warnings.catch_warnings():
warnings.filterwarnings('ignore')
# fit dist to data
params = distribution.fit(self.data)
# Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
# Calculate fitted PDF and error with fit in distribution
pdf = distribution.pdf(x, loc=loc, scale=scale, *arg)
sse = np.sum(np.power(y - pdf, 2.0))
# if axis pass in add to plot
try:
if ax:
pd.Series(pdf, x).plot(ax=ax)
except Exception:
pass
# identify if this distribution is better
best_distributions.append((distribution, params, sse))
except Exception as ex:
if self.debug:
trace=traceback.format_exc()
msg=f"fit for {distributionName} failed:{ex}
{trace}"
print(msg,file=sys.stderr)
pass
return sorted(best_distributions, key=lambda x:x[2])
def make_pdf(self,dist, params:list, size=10000):
"""
Generate distributions's Probability Distribution Function
Args:
dist: Distribution
params(list): parameter
size(int): size
Returns:
dataframe: Power Distribution Function
"""
# Separate parts of parameters
arg = params[:-2]
loc = params[-2]
scale = params[-1]
# Get sane start and end points of distribution
start = dist.ppf(0.01, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.01, loc=loc, scale=scale)
end = dist.ppf(0.99, *arg, loc=loc, scale=scale) if arg else dist.ppf(0.99, loc=loc, scale=scale)
# Build PDF and turn into pandas Series
x = np.linspace(start, end, size)
y = dist.pdf(x, loc=loc, scale=scale, *arg)
pdf = pd.Series(y, x)
return pdf
def analyze(self,title,x_label,y_label,outputFilePrefix=None,imageFormat:str='png',allBins:int=50,distBins:int=200,density:bool=True):
"""
analyze the Probabilty Distribution Function
Args:
data: Panda Dataframe or numpy array
title(str): the title to use
x_label(str): the label for the x-axis
y_label(str): the label for the y-axis
outputFilePrefix(str): the prefix of the outputFile
imageFormat(str): imageFormat e.g. png,svg
allBins(int): the number of bins for all
distBins(int): the number of bins for the distribution
density(bool): if True show relative density
"""
self.allBins=allBins
self.distBins=distBins
self.density=density
self.title=title
self.x_label=x_label
self.y_label=y_label
self.imageFormat=imageFormat
self.outputFilePrefix=outputFilePrefix
self.color=list(matplotlib.rcParams['axes.prop_cycle'])[1]['color']
self.best_dist=None
self.analyzeAll()
if outputFilePrefix is not None:
self.saveFig(f"{outputFilePrefix}All.{imageFormat}", imageFormat)
plt.close(self.figAll)
if self.best_dist:
self.analyzeBest()
if outputFilePrefix is not None:
self.saveFig(f"{outputFilePrefix}Best.{imageFormat}", imageFormat)
plt.close(self.figBest)
def analyzeAll(self):
'''
analyze the given data
'''
# Plot for comparison
figTitle=f"{self.title}
All Fitted Distributions"
self.figAll=plt.figure(figTitle,figsize=(12,8))
ax = self.data.plot(kind='hist', bins=self.allBins, density=self.density, alpha=0.5, color=self.color)
# Save plot limits
dataYLim = ax.get_ylim()
# Update plots
ax.set_ylim(dataYLim)
ax.set_title(figTitle)
ax.set_xlabel(self.x_label)
ax.set_ylabel(self.y_label)
# Find best fit distribution
best_distributions = self.best_fit_distribution(bins=self.distBins, ax=ax,density=self.density)
if len(best_distributions)>0:
self.best_dist = best_distributions[0]
# Make PDF with best params
self.pdf = self.make_pdf(self.best_dist[0], self.best_dist[1])
def analyzeBest(self):
'''
analyze the Best Property Distribution function
'''
# Display
figLabel="PDF"
self.figBest=plt.figure(figLabel,figsize=(12,8))
ax = self.pdf.plot(lw=2, label=figLabel, legend=True)
self.data.plot(kind='hist', bins=self.allBins, density=self.density, alpha=0.5, label='Data', legend=True, ax=ax,color=self.color)
param_names = (self.best_dist[0].shapes + ', loc, scale').split(', ') if self.best_dist[0].shapes else ['loc', 'scale']
param_str = ', '.join(['{}={:0.2f}'.format(k,v) for k,v in zip(param_names, self.best_dist[1])])
dist_str = '{}({})'.format(self.best_dist[0].name, param_str)
ax.set_title(f'{self.title} with best fit distribution
' + dist_str)
ax.set_xlabel(self.x_label)
ax.set_ylabel(self.y_label)
def saveFig(self,outputFile:str=None,imageFormat='png'):
'''
save the current Figure to the given outputFile
Args:
outputFile(str): the outputFile to save to
imageFormat(str): the imageFormat to use e.g. png/svg
'''
plt.savefig(outputFile, format=imageFormat) # dpi
if __name__ == '__main__':
# Load data from statsmodels datasets
data = pd.Series(sm.datasets.elnino.load_pandas().data.set_index('YEAR').values.ravel())
bfd=BestFitDistribution(data)
for density in [True,False]:
suffix="density" if density else ""
bfd.analyze(title=u'El Niño sea temp.',x_label=u'Temp (°C)',y_label='Frequency',outputFilePrefix=f"/tmp/ElNinoPDF{suffix}",density=density)
# uncomment for interactive display
# plt.show()







